How Infopulse Took Part in Kaggle Data Science Bowl 2017
About Kaggle Platform
Kaggle is the largest platform for data scientists, hosting a series of online competitions with open data. Sponsored and organized by world’s largest companies like Google, Intel, Mercedes-Benz, MasterCard, Amazon, NVidia and others, Kaggle has become a sort of Olympic Games for the best data science teams worldwide. The main task of Kaggle Challenge is to design working software models using real data. The model codes can later be accepted for production to improve existing solutions or to build new ones. While the prizes in the competition can reach hundreds of thousands of USD, participation itself is a huge achievement for any team.
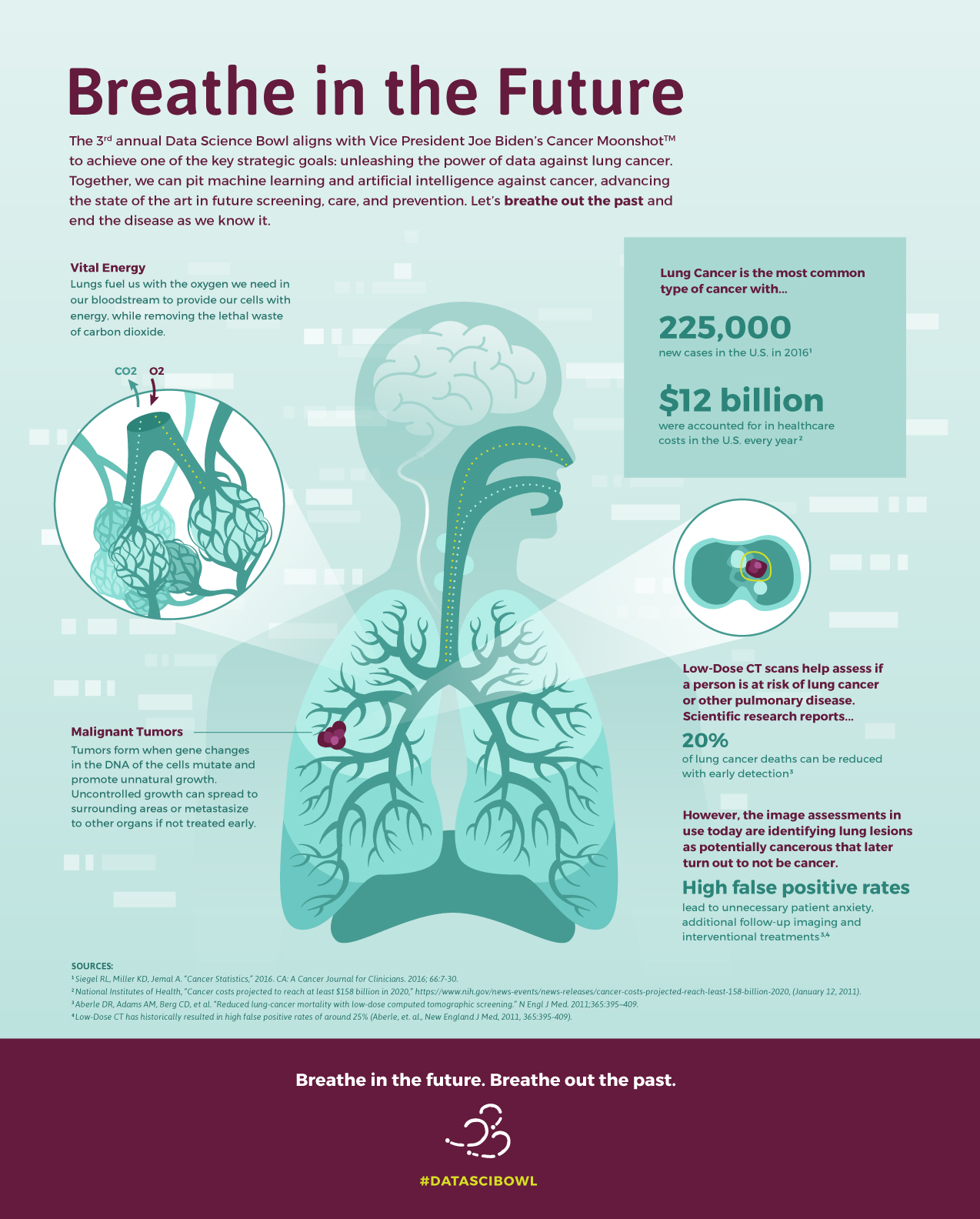
Organized by the National Science Academy, Kaggle Data Science Bowl 2017 has become one of the largest competitions in the history of Kaggle, with the prize fund totaling $1Mln. During 90 days of the event, participants were required to design working models that would check the CT scans of lungs for cancer. The ultimate aim of the competition was to create cancer screening and prevention algorithms that would help increase the accuracy of cancerous lesions detection and improve patients’ treatment.
Oleg Nalyvaiko, Infopulse Head of cognitive computing department, gathered a team of data science experts and machine learning enthusiasts to participate in the event. The Infopulse team was among the few teams from Ukraine, who took part in the competition.
About the challenge
Kaggle Data Science Bowl 2017 had two stages. During the initial first stage, we received access to AWS training stage and a huge pile of data – a 150GB set of CT images belonging over 1,500 patients. While all teams had enough time to design and trial-test their models during the first stage, closer to the end of the competition organizers initiated the second stage to make sure the participants were not cheating. A new set of data was published, with a new test dataset for model evaluation, extending the total dataset to almost 2,000 patients. Only 5 days were given for processing of the second part of data. Thus, only real models remained by the end. Along with the answers to the challenge, the teams were asked to upload the source code of their models as well for additional verification on Kaggle side.
Despite lacking a dedicated biologist in our team or high-level medical skills, Infopulse team was ready to embrace the challenge. This is how we worked.
Data processing and learning model
According to the task, we needed to demonstrate the rating of data and preciseness of our model according to the logarithmic deviation. We had to go through many discussions regarding means and principles of data processing and analysis, as incoming data was in a DICOM format, used for CT and MRI imaging. A patient case contained approximately two hundreds of 2D frames with a certain possibility to have cancer.
For starters, we needed to convert this data format so that we could work with it. We trial-tested several approaches while looking for the best one to accomplish this task.
The first idea was to build a 3D Convolutional neural network and train it with provided data. What about scanning? The unit of measurement in CT scans is the Hounsfield Unit (HU – a measure of radiodensity). CT scanners are carefully calibrated to accurately measure this. We filtered all redundant info (air, bones, etc.) from the images according to the Hounsfield scale. Then we resampled the full dataset to a certain isotropic resolution.
And finally, when we trained our network on a simple 3D CNN and checked it’s accuracy with CV data, we found that the results were almost the same as naïve submissions, filled with 0.5 values. We decided to discard the 3D approach, as it wasn’t a good practice for this task – well, at least due to the limited amount of data available.
Next, we tried another approach, recommended by other participants – to use a pre-trained ResNet50 model as features extractor. We reduced the dimensionality of obtained data with PCA and then fed it to XGB-classifier. The result was much better and we got to the Top 10% with this approach on the Public Leaderboard during the 1st stage of the competition. At the beginning of the 2nd stage, we were placed on the 2nd place. However, after everyone made their final submission, we went down to the 79th place. The Kaggle Leaderboard system is tricky, and after publishing the final Private Leaderboard, we were placed 278 out of almost 2000 submissions with this model, which showed that it was strongly over-fitted.
Our last approach was based on LUNA16 competition 2016 results. This competition was based on Lungs Nodule Analysis (hence LUNA) and provided lungs images with labeled nodules. We used this data to train our model, which would then allow locating zones of interest on our images. U-net CNN neural network was the best fit for this task, as it was created specifically for the biomedical image segmentation. After training our segmentation model, we applied a DSB dataset. The obtained results were fed to the XGB-classifier. This allowed us to receive a log-loss result on cross-validation of approximately 0.4. Unfortunately, we didn’t have enough time to conduct the exhaustive computations and make the final submission on time with this latest model. We started implementing this idea closer to the end of the competition and didn’t expect that such operation as image preprocessing (LUNA16 and DSB 2017) would be this time-consuming. Approximately 70 hours were spent on image preprocessing and 2 days to train a U-net. We’re talking about pure computing time, without code writing or debugging. Still, we consider this to be our best and well thought-out approach.

We’ve written our code with Python, and used Keras with TensorFlow back-end to build a deep learning framework.
All computations were performed on a local machine:
- Intel Core i7-6700k (overclocked to 4.5 GHz)
- 16 GB of RAM (2666 MHz)
- NVidia GeForce GTX 980 Ti (with frequencies boost)
- Ubuntu 16.04, Python 3, TensorFlow 1.0
What’s next?
The competition was really tough! While we reached the second place in the mid-way, we ended up on a 278th place out of almost 2000 teams in the end. We still gained a great experience and skills, proving to ourselves that nothing is impossible. Our project source code was uploaded in a simple form to our GitHub repository.
In the meantime, our team is super-enthusiastic about participating in other events in the future and doing new data science projects.
As a reminder, recently Infopulse launched a dedicated Cognitive Computing department, focused on data science and related areas, and right now we’re working on chatbots for ITSM.

![CX with Virtual Assistants in Telecom [thumbnail]](/uploads/media/280x222-how-to-improve-cx-in-telecom-with-virtual-assistants.webp)
![Generative AI and Power BI [thumbnail]](/uploads/media/thumbnail-280x222-generative-AI-and-Power-BI-a-powerful.webp)
![AI for Risk Assessment in Insurance [thumbnail]](/uploads/media/aI-enabled-risk-assessment_280x222.webp)
![Super Apps Review [thumbnail]](/uploads/media/thumbnail-280x222-introducing-Super-App-a-Better-Approach-to-All-in-One-Experience.webp)
![IoT Energy Management Solutions [thumbnail]](/uploads/media/thumbnail-280x222-iot-energy-management-benefits-use-сases-and-сhallenges.webp)
![5G Network Holes [Thumbnail]](/uploads/media/280x222-how-to-detect-and-predict-5g-network-coverage-holes.webp)

![How to Reduce Churn in Telecom [thumbnail]](/uploads/media/thumbnail-280x222-how-to-reduce-churn-in-telecom-6-practical-strategies-for-telco-managers.webp)
![Automated Machine Data Collection for Manufacturing [Thumbnail]](/uploads/media/thumbnail-280x222-how-to-set-up-automated-machine-data-collection-for-manufacturing.webp)
![Money20/20 Key Points [thumbnail]](/uploads/media/thumbnail-280x222-humanizing-the-fintech-industry-money-20-20-takeaways.webp)